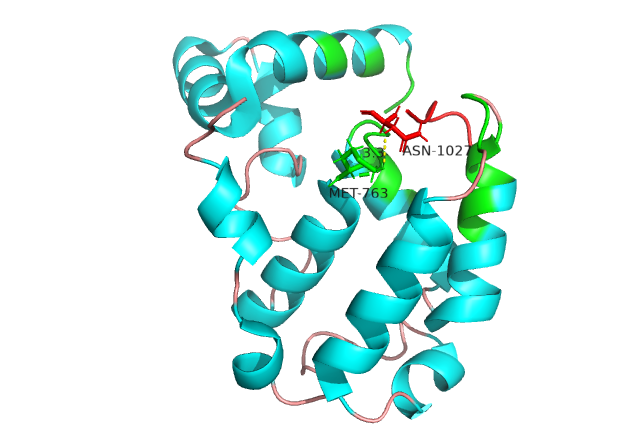
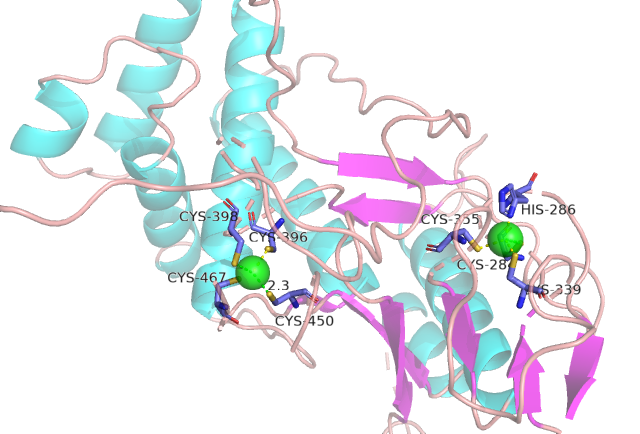
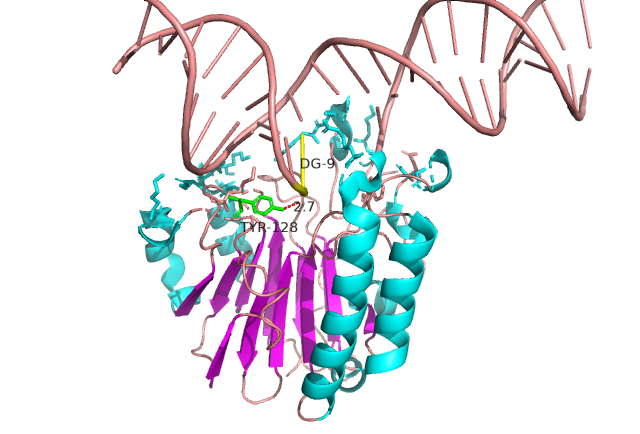
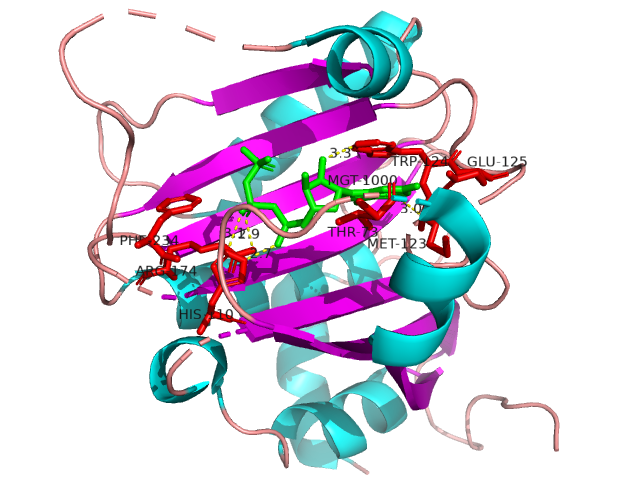
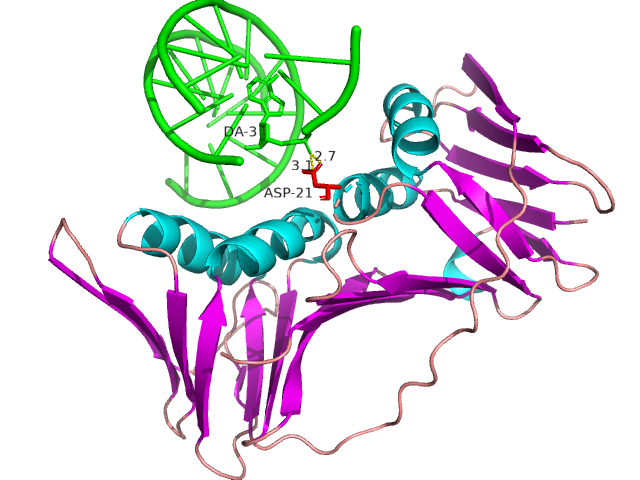
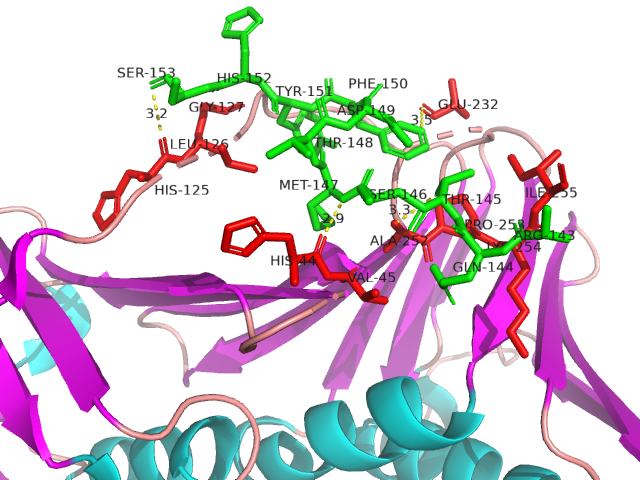
IDPsBind is a repository of binding sites for IDPs complexes with known 3D structure. IDPsBind uses a
distance cutoff 3.5 angstroms to identify which amino acids contact ligand in solved complex structures from
the PDB database and annotates these interaction binding sites. IDPsBind contains 9626 complex structures in
880 entries. 880 intrinsically disordered proteins are selected from the Disprot database. And IDPsBind
contains 3203 binding ligands.
IDPsBind: 1.0
Release: 2021_07